In the fields of molecular biology and genetics, a genome is the genetic material of an organism. It consists of DNA (or RNA in RNA viruses). Each genome contains all of the information needed to build and maintain that organism. In humans, a copy of the entire genome—more than 3 billion DNA base pairs—is contained in all cells that have a nucleus.
In bioinformatics, a genome browser is a graphical interface for display of information from a biological database for genomic data. They are important tools for studying genomes given the vast amounts of data available. They typically load very large files, such as whole genome FASTA files and display them in a way that users can make sense of the information there. They can be used to visualize a variety of different data types.
Genome browsers enable researchers to visualize and browse entire genomes with annotated data including gene prediction and structure, proteins, expression, regulation, variation, comparative analysis, etc. They use a visual, high-level overview of complex data in a form that can be grasped at a glance and provide the means to explore the data in increasing resolution from megabase scales down to the level of individual elements of the DNA sequence.
There are numerous tools to browse genomes. While there’s considerable overlap in functionality, each software offers unique features or data. Most genome browsers don’t require any programming knowledge.
Here are our recommendations summarized in the chart below. This article focuses solely on standalone desktop genome browsers. Many of them use Java and so are cross-platform tools.
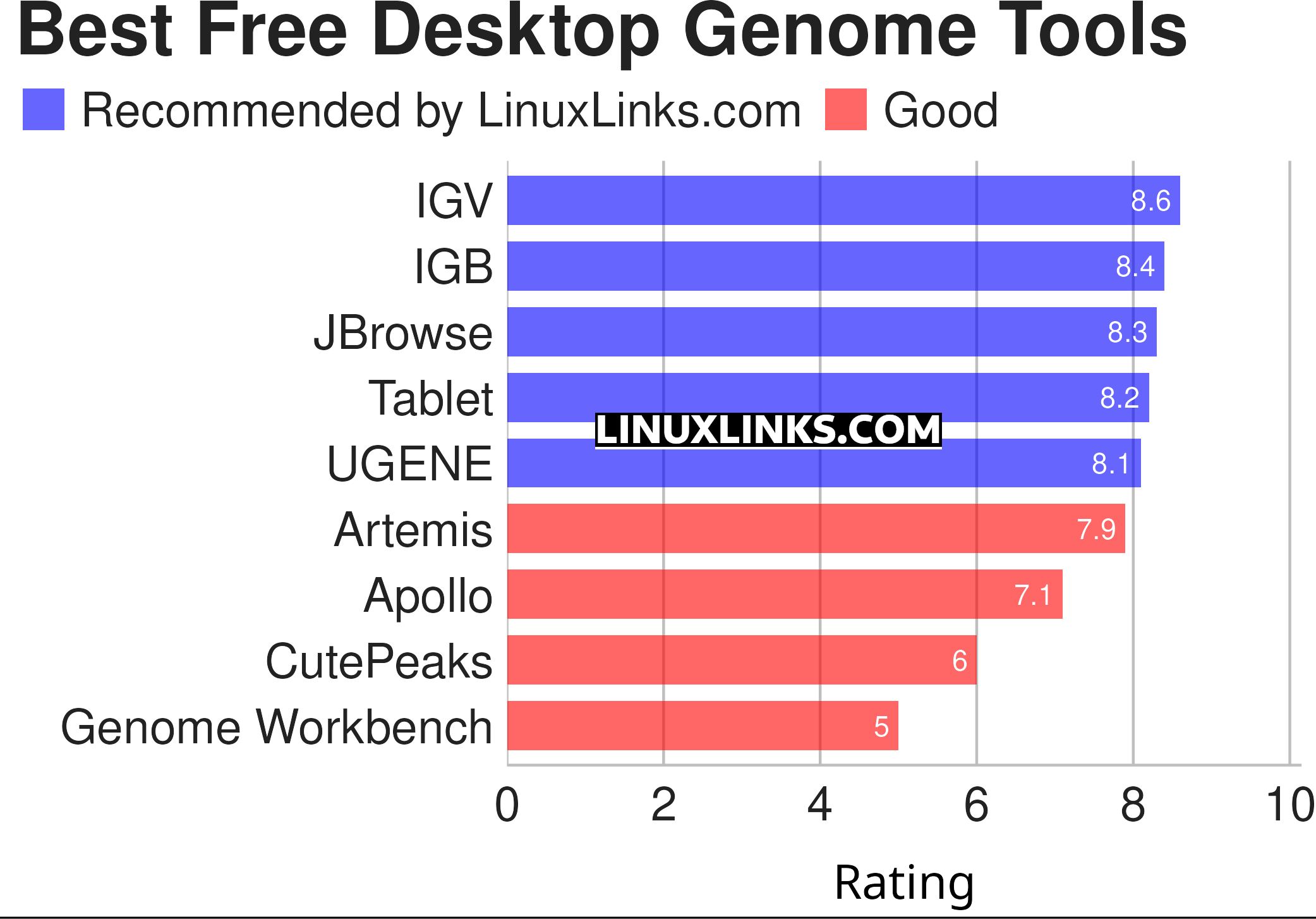
For more details about each application, we’ve compiled dedicated pages below detailing their features, a screenshot, and links to useful resources.
| Desktop Genome Browsers | |
|---|---|
| IGV | High-performance visualization genome browser tool |
| IGB | Integrated Genome Browser |
| JBrowse | Fast, scalable genome browser |
| Tablet | Graphical viewer for sequence assemblies and alignments |
| UGENE | Set of integrated bioinformatics software |
| Artemis | Genome viewer and annotation tool |
| Apollo | Instantaneous, collaborative genomic annotation editor |
| CutePeaks | Cross platform Sanger Trace file viewer |
| Genome Workbench | Integrated tools for studying and analyzing genetic data |
Note: This Group Test has deliberately excluded web based genome browsers. These are covered separately in this roundup.
 Read our complete collection of recommended free and open source software. Our curated compilation covers all categories of software. Read our complete collection of recommended free and open source software. Our curated compilation covers all categories of software. Spotted a useful open source Linux program not covered on our site? Please let us know by completing this form. The software collection forms part of our series of informative articles for Linux enthusiasts. There are hundreds of in-depth reviews, open source alternatives to proprietary software from large corporations like Google, Microsoft, Apple, Adobe, IBM, Cisco, Oracle, and Autodesk. There are also fun things to try, hardware, free programming books and tutorials, and much more. |

