geWorkbench (genomics Workbench) is a Java-based open-source platform for integrated genomics. Using a component architecture it allows individually developed plug-ins to be configured into complex bioinformatic applications.
At present there are more than 70 available plug-ins supporting the visualization and analysis of gene expression and sequence data.
geWorkbench is the Bioinformatics platform of MAGNet, the National Center for the Multi-scale Analysis of Genomic and Cellular Networks, one of the 8 National Centers for Biomedical Computing.
Features include:
- Computational analysis tools such as t-test, hierarchical clustering, self-organizing maps, regulatory network reconstruction, BLAST searches, pattern-motif discovery, protein structure prediction, structure-based protein annotation, etc.
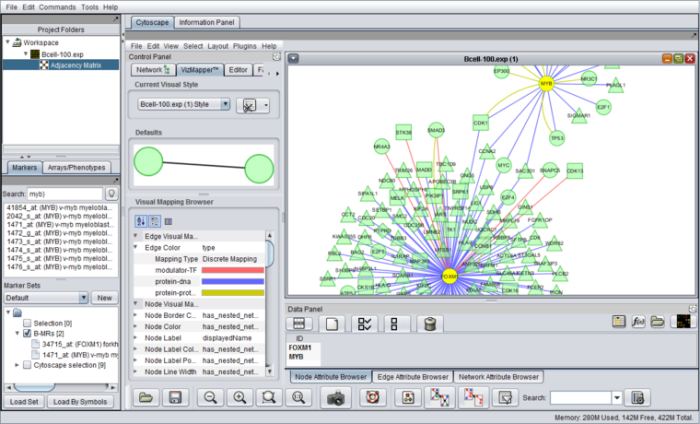
- Visualization of gene expression (heatmaps, volcano plot), molecular interaction networks (through Cytoscape), protein sequence and protein structure data (e.g., MarkUs).
- Integration of gene and pathway annotation information from curated sources as well as through Gene Ontology enrichment analysis.
- Component integration through platform management of inputs and outputs. Among data that can be shared between components are expression datasets, interaction networks, sample and marker (gene) sets and sequences.
- Dataset history tracking – complete record of data sets used and input settings.
- Integration with 3rd party tools such as Genepattern, Cytoscape, and Genomespace.
- Provides an environment which supports moving from one data type to another in a seamless fashion, e.g. from gene expression to sequences to patterns.
- Provides access to a variety of external data sources, including:
- Microarray gene expression repositories (caArray).
- BLAST (NCBI).
- Gene annotation pages (via bioDBNet).
- Protein and DNA sequence retrieval (UC Santa Cruz and EBI).
- Pathway diagrams (BioCarta).
- Provides a gateway to several computational services currently hosted on Columbia servers and clusters, including:
-
- Pattern Discovery.
- Pudge – protein structure modeling.
- SkyBase – database of molecular models.
-
Specific types of data supported include:
- Microarray Gene Expression:
- GEO Soft: Series, Series Matrix, and Annotated Matrix (GDS).
- MAGE-TAB data matrix.
- Affymetrix GCOS/MAS5.
- Matrix format (geWorkbench).
- Tab-delimited (e.g. RMAExpress).
- GenePix.
- Microarray Gene Expression Annotation file support:
- Affymetrix 3′ Expression.
- Affymetrix WT Gene/Exon ST (transcript-level) including Gene Array 1.0/2.0 ST and Exon 1.0 ST.
- DNA and Protein Sequences:
- FASTA.
- Pathways:
- BioCarta.
- Molecular structure – prediction, annotation and display.
- Sequence Patterns:
- Regular Expressions.
- Gene Ontology.
- Regulatory Networks.
Website: github.com/floratos-lab/geworkbench-core
Support:
Developer: Columbia University, First Genetic Trust National Cancer Institute
License: BSD-like
geWorkbench is written in Java. Learn Java with our recommended free books and free tutorials.
Return to Bioinformatics Tools
| Popular series | |
|---|---|
| The largest compilation of the best free and open source software in the universe. Each article is supplied with a legendary ratings chart helping you to make informed decisions. | |
| Hundreds of in-depth reviews offering our unbiased and expert opinion on software. We offer helpful and impartial information. | |
| The Big List of Active Linux Distros is a large compilation of actively developed Linux distributions. | |
| Replace proprietary software with open source alternatives: Google, Microsoft, Apple, Adobe, IBM, Autodesk, Oracle, Atlassian, Corel, Cisco, Intuit, SAS, Progress, Salesforce, and Citrix | |
| Awesome Free Linux Games Tools showcases a series of tools that making gaming on Linux a more pleasurable experience. This is a new series. | |
| Machine Learning explores practical applications of machine learning and deep learning from a Linux perspective. We've written reviews of more than 40 self-hosted apps. All are free and open source. | |
| New to Linux? Read our Linux for Starters series. We start right at the basics and teach you everything you need to know to get started with Linux. | |
| Alternatives to popular CLI tools showcases essential tools that are modern replacements for core Linux utilities. | |
| Essential Linux system tools focuses on small, indispensable utilities, useful for system administrators as well as regular users. | |
| Linux utilities to maximise your productivity. Small, indispensable tools, useful for anyone running a Linux machine. | |
| Surveys popular streaming services from a Linux perspective: Amazon Music Unlimited, Myuzi, Spotify, Deezer, Tidal. | |
| Saving Money with Linux looks at how you can reduce your energy bills running Linux. | |
| Home computers became commonplace in the 1980s. Emulate home computers including the Commodore 64, Amiga, Atari ST, ZX81, Amstrad CPC, and ZX Spectrum. | |
| Now and Then examines how promising open source software fared over the years. It can be a bumpy ride. | |
| Linux at Home looks at a range of home activities where Linux can play its part, making the most of our time at home, keeping active and engaged. | |
| Linux Candy reveals the lighter side of Linux. Have some fun and escape from the daily drudgery. | |
| Getting Started with Docker helps you master Docker, a set of platform as a service products that delivers software in packages called containers. | |
| Best Free Android Apps. We showcase free Android apps that are definitely worth downloading. There's a strict eligibility criteria for inclusion in this series. | |
| These best free books accelerate your learning of every programming language. Learn a new language today! | |
| These free tutorials offer the perfect tonic to our free programming books series. | |
| Linux Around The World showcases usergroups that are relevant to Linux enthusiasts. Great ways to meet up with fellow enthusiasts. | |
| Stars and Stripes is an occasional series looking at the impact of Linux in the USA. | |

